import numpy as np
from matplotlib import pyplot
import skimage as skiscore
Pixel-wise H score
compute_pxlhscore
compute_pxlhscore (hed_img, h_threshold=0.05, d_thresholds=[0.12, 0.24, 0.6], verbose=False)
*Computes the pixel H-score for a given HED (Hematoxylin and Eosin-DAB) stained image.
The H-score is calculated based on the intensity of the DAB stain, which is indicative of the presence and quantity of a specific biomarker in IHC images. The function allows for automatic thresholding based on the distribution of staining intensities. Inspired by the implementation in Ram et al. 2021.
Parameters:
hed_img (numpy.ndarray): The HED-stained image as a NumPy array of shape (height, width, channels).
h_threshold (str or float): Threshold for Hematoxylin intensity. If ‘default’, the threshold is set to 0.05. If ‘static’, the threshold is set to the mean intensity. If ‘auto’, the threshold is set based on Otsu’s thresholding.
d_thresholds (str or list of floats): Thresholds for DAB intensity, defining negative, low, medium, and high intensity ranges. If ‘default’, thresholds are set to [0.12, 0.24, 0.6]. If ‘static’, thresholds are set to the 90th, 94.95th, and 99.9th percentiles. If ‘auto’, thresholds are set based on Otsu’s thresholding for 3 classes.
verbose (bool): If True, displays histograms of the distributions of Hematoxylin and DAB stain values, and images showing pixels classified as high, medium, low, and negative DAB stained.
Returns:
- pxlHscore (float): The pixel H-score, a weighted sum of pixels classified as having high, medium, or low DAB intensity, normalized by the total number of pixels considered.*
The pixelwise H-score is computed as follows:
\[\ pxlH = 100 \times \frac{3 \times N_{DH} + 2 \times N_{DM} + N_{DL}}{N_{DH} + N_{DM} + N_{DL} + N_{N}} \]
where:
\(\N_{DH}\) is the number of pixels with high DAB intensity,
\(\N_{DM}\) is the number of pixels with medium DAB intensity,
\(\N_{DL}\) is the number of pixels with low DAB intensity,
\(\N_{N}\) is the number of pixels with negative DAB intensity.
Example
ihc_rgb = ski.data.immunohistochemistry()
pyplot.imshow(ihc_rgb)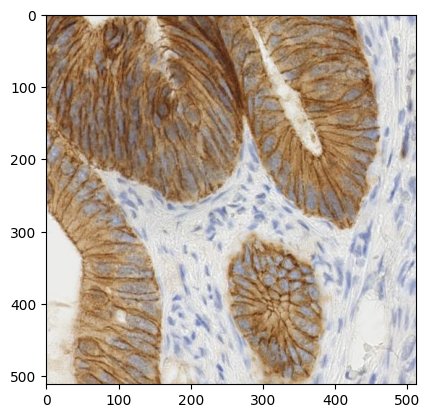
ihc_hed = ski.color.rgb2hed(ihc_rgb)stain_names = ["Hematoxylin", "Eosin", "DAB"]
fig, axes = pyplot.subplots(1, 3, figsize=(12, 6))
null = np.zeros_like(ihc_hed[:, :, 0])
for i, name in enumerate(stain_names):
# Create an RGB image for the current stain
channels = [null, null, null]
channels[i] = ihc_hed[:, :, i]
rgb_img = ski.color.hed2rgb(np.stack(channels, axis=-1))
# Display
axes[i].imshow(rgb_img)
axes[i].set_title(name)
axes[i].axis("off")
pyplot.tight_layout()
pyplot.show()
result = compute_pxlhscore(ihc_hed, verbose=True)
print(result)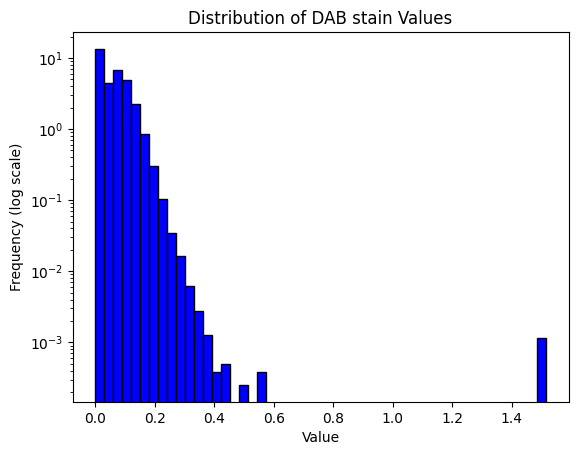
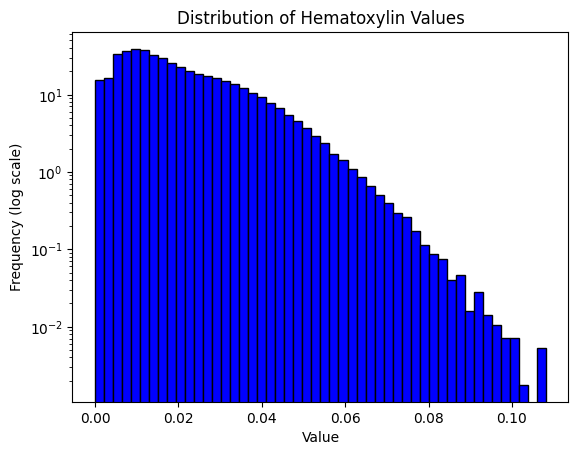
Mean intensity for H pixels: 0.035449981689453125
Mean intensity for DN pixels: 0.88824462890625
Mean intensity for DL pixels: 0.10962677001953125
Mean intensity for DM pixels: 0.002094268798828125
Mean intensity for DH pixels: 3.4332275390625e-05
Mean intensity for N pixels: 0.031993865966796875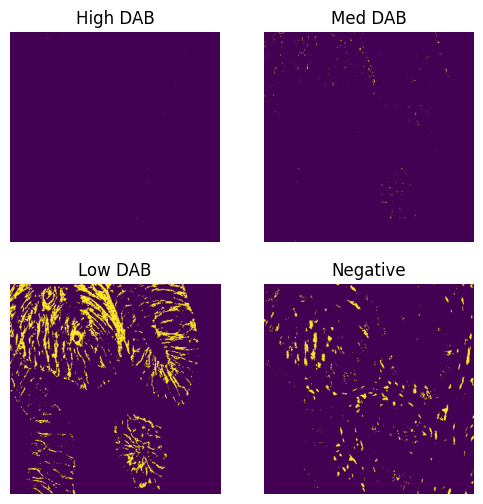
Haematoxylin threshold: 0.05
DAB thresholds: [0.12, 0.24, 0.6]
Pixel H-score: 79.24793673539793
79.24793673539793assert result == compute_pxlhscore(ihc_hed, d_thresholds=[0.12, 0.24, 0.6]) == compute_pxlhscore(ihc_hed, d_thresholds="default")assert result == compute_pxlhscore(ihc_hed, h_threshold=0.05) == compute_pxlhscore(ihc_hed, h_threshold="default")result = compute_pxlhscore(ihc_hed, h_threshold="static", d_thresholds="static", verbose=True)
print(result)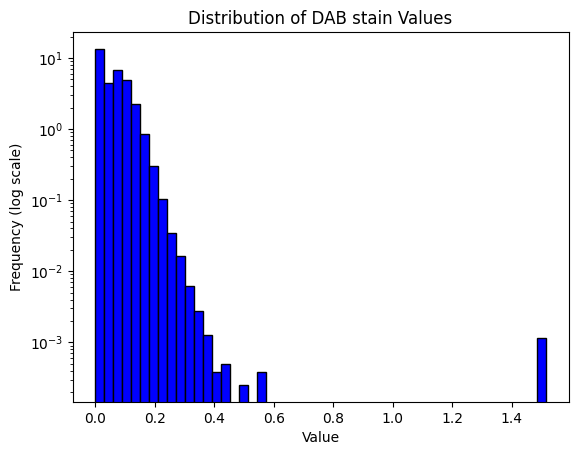
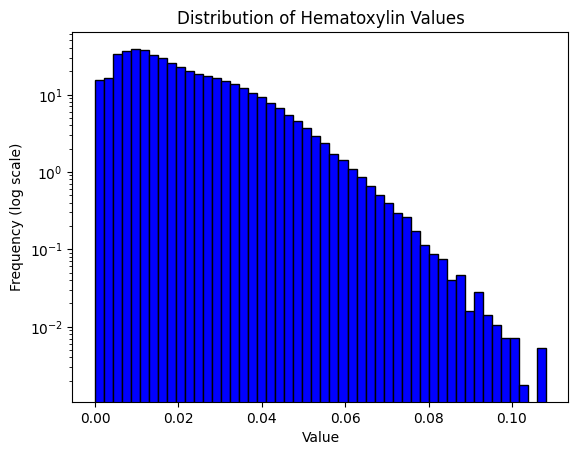
Mean intensity for H pixels: 0.4108695983886719
Mean intensity for DN pixels: 0.8999786376953125
Mean intensity for DL pixels: 0.04950714111328125
Mean intensity for DM pixels: 0.049510955810546875
Mean intensity for DH pixels: 0.001003265380859375
Mean intensity for N pixels: 0.3635520935058594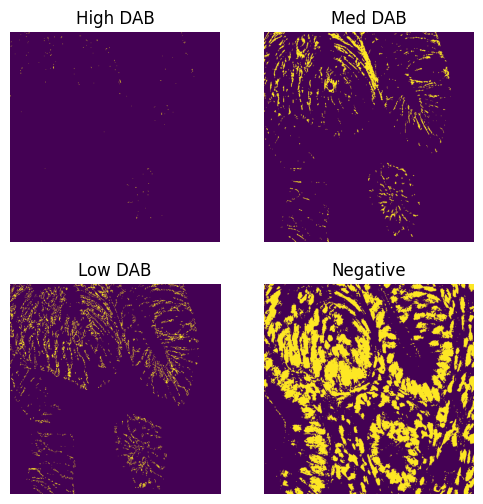
Haematoxylin threshold: 0.02006582564164016
DAB thresholds: [0.12352821 0.14450278 0.26666182]
Pixel H-score: 32.689285155896414
32.689285155896414result = compute_pxlhscore(ihc_hed, h_threshold="auto", d_thresholds="auto", verbose=True)
print(result)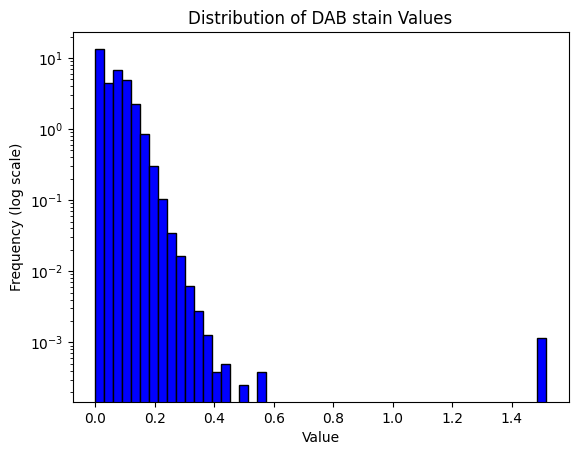
Mean intensity for H pixels: 0.3234710693359375
Mean intensity for DN pixels: 0.43198394775390625
Mean intensity for DL pixels: 0.279510498046875
Mean intensity for DM pixels: 0.22780609130859375
Mean intensity for DH pixels: 0.060699462890625
Mean intensity for N pixels: 0.10207366943359375
Haematoxylin threshold: 0.024294931878818742
DAB thresholds: [0.03847427 0.08582721 0.13909927]
Pixel H-score: 136.88033701468746
136.88033701468746